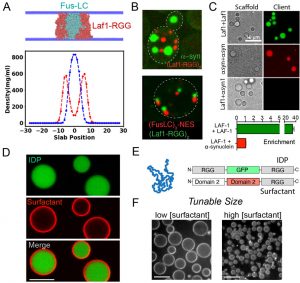
Figure 1. A. CG co-simulations identifies IDPs that selectively coacervate. B. Formation of distinct compartments from IDPs co-expressed in cells. C. Chemical reconstitution shows specificity encoded directly in polypeptide chain. D. Composite condensates with surface active ‘skins’. E. Modular block strategy for protein surfactants. F. Control of condensate size using surfactants
Using coarse-grain simulations (1) and biochemical reconstitution, our team identified intrinsically disordered proteins that do not mix with the Laf-1 RGG polypeptide and instead form distinct mesoscale condensate materials. Good and Rhoades characterized these polymer sequences in vitro and through co-expression in cells, demonstrating that coacervation specificity is hard-coded in the polypeptide chain. Hammer, Good and Mittal identified amino acid sequence features that drive Laf-1 RGG condensation (2) and are extending this approach to characterize the sequence determinants of coacervation specificity.
Additionally, our team has developed surface active agents built from protein blocks that localize to condensate interfaces and enable tunable control of condensate size. Hammer and Lee are now using this platform generate skins that control the liquid properties of condensates and to assemble collectives of coacervates into granular materials.
1Sequence dependent phase separation of protein-polynucleotide mixtures elucidated using molecular simulations. R. M. Regy, G. L. Dignon, W. Zheng, Y. C. Kim, J. Mittal. NAR, 48, 12593-12603 (2020).
2Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Schuster BS, Dignon GL, Tang WS, Kelley FM, Ranganath AK, Jahnke CN, Simpkins AG, Regy RM, Hammer DA, Good MC, Mittal J. PNAS 117, 11421-11431 (2020).
